Jupyter Snippet CB2nd 01_markov
Jupyter Snippet CB2nd 01_markov
13.1. Simulating a discrete-time Markov chain
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
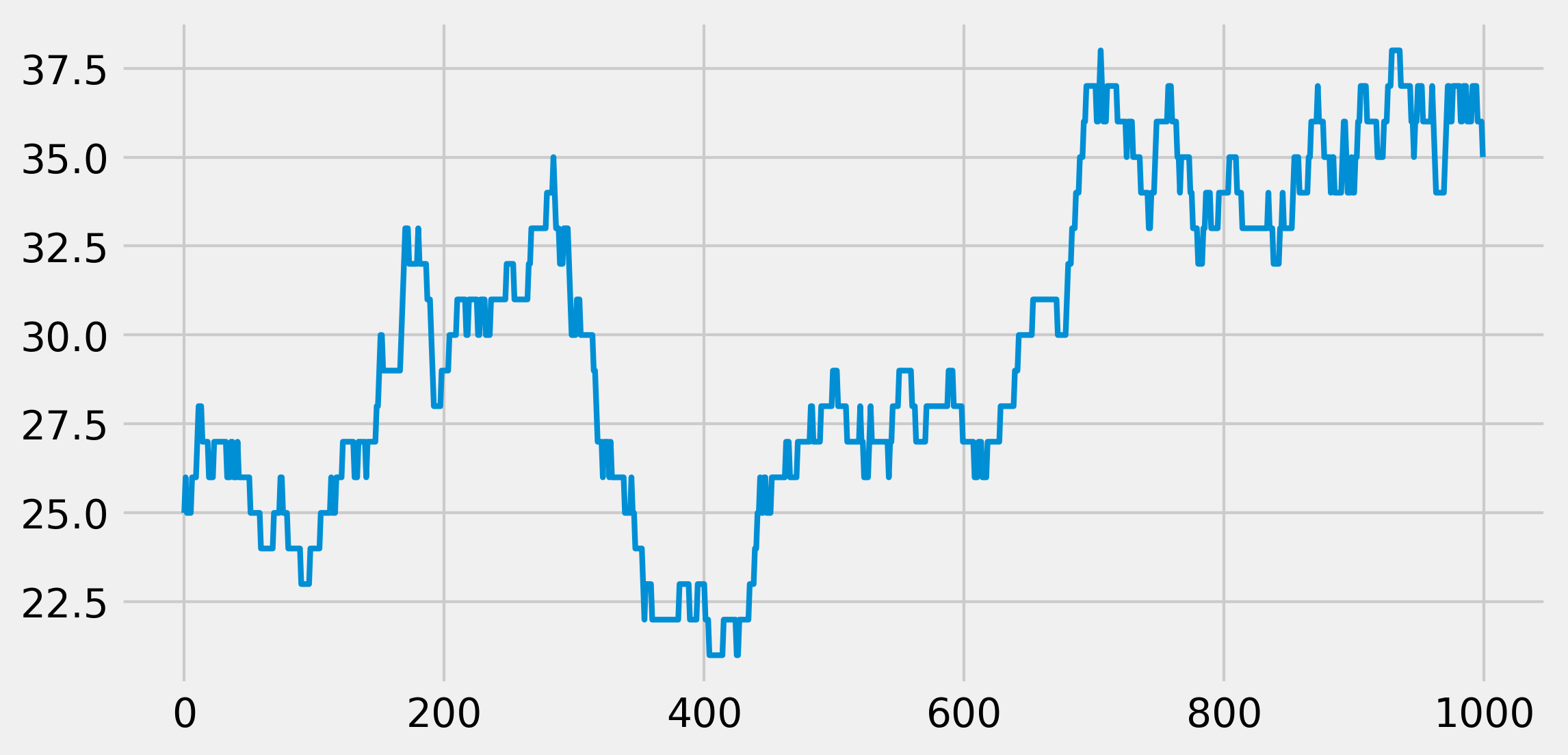
N = 100 # maximum population size
a = .5 / N # birth rate
b = .5 / N # death rate
nsteps = 1000
x = np.zeros(nsteps)
x[0] = 25
for t in range(nsteps - 1):
if 0 < x[t] < N - 1:
# Is there a birth?
birth = np.random.rand() <= a * x[t]
# Is there a death?
death = np.random.rand() <= b * x[t]
# We update the population size.
x[t + 1] = x[t] + 1 * birth - 1 * death
# The evolution stops if we reach $0$ or $N$.
else:
x[t + 1] = x[t]
fig, ax = plt.subplots(1, 1, figsize=(8, 4))
ax.plot(x, lw=2)
ntrials = 100
x = np.random.randint(size=ntrials,
low=0, high=N)
def simulate(x, nsteps):
"""Run the simulation."""
for _ in range(nsteps - 1):
# Which trials to update?
upd = (0 < x) & (x < N - 1)
# In which trials do births occur?
birth = 1 * (np.random.rand(ntrials) <= a * x)
# In which trials do deaths occur?
death = 1 * (np.random.rand(ntrials) <= b * x)
# We update the population size for all trials
x[upd] += birth[upd] - death[upd]
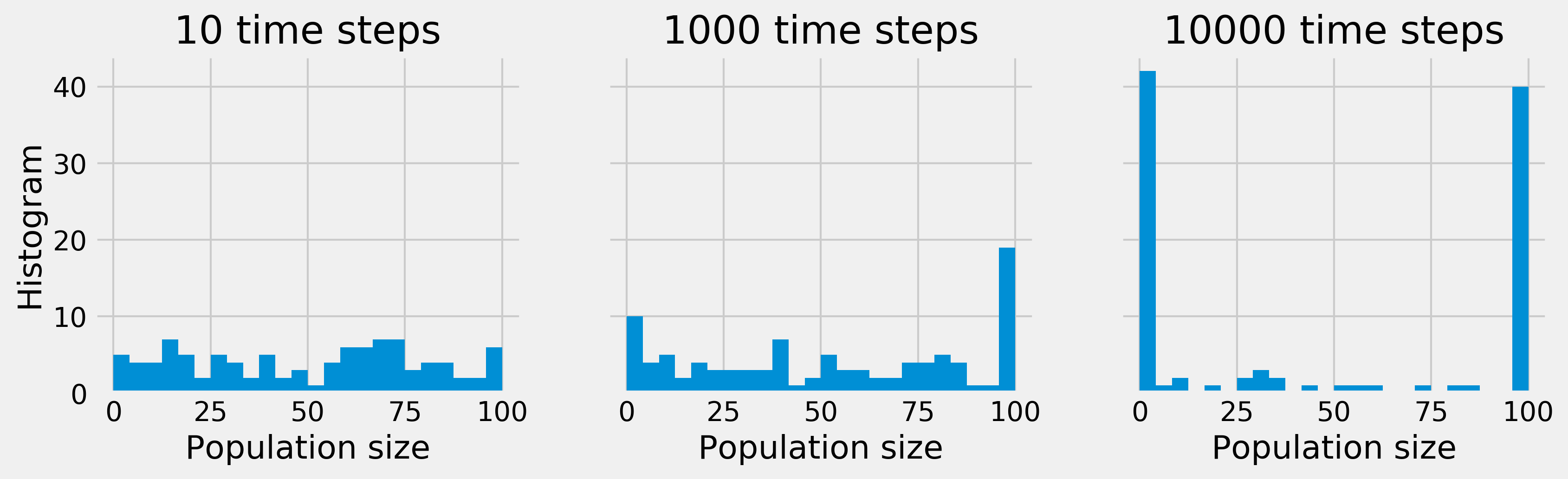
bins = np.linspace(0, N, 25)
nsteps_list = [10, 1000, 10000]
fig, axes = plt.subplots(1, len(nsteps_list),
figsize=(12, 3),
sharey=True)
for i, nsteps in enumerate(nsteps_list):
ax = axes[i]
simulate(x, nsteps)
ax.hist(x, bins=bins)
ax.set_xlabel("Population size")
if i == 0:
ax.set_ylabel("Histogram")
ax.set_title(f"{nsteps} time steps")